Phylogenomics and Molecular Evolution
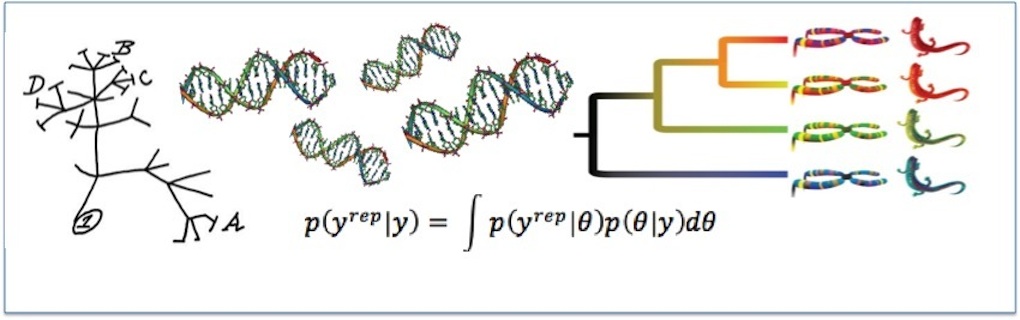
Our research focuses on the intersection of genomics, phylogenetics, and molecular evolution. We are interested in how to best use genomic sequencies to infer evolutionary history and also understand how evolution has shaped the genome itself.
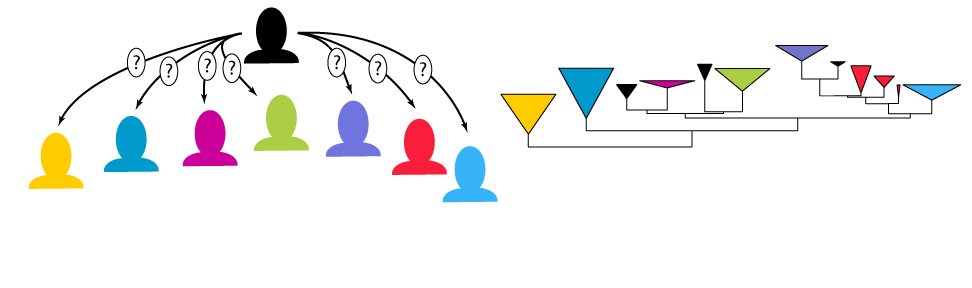
Why phylogenies?
Evolutionary biologists are driven by a desire to understand the staggering diversity of life on Earth, its history of change through time and the processes that have driven such change. DNA sequences are the fundamental material upon which evolutionary change is wrought and each organism’s genome carries the legacies of its evolutionary history. By sequencing the genomes of different organisms, we can use the information they contain to reconstruct their historical relationships and test hypotheses about their evolutionary past. Organismal histories can be depicted as branching diagrams known as phylogenetic trees. Phylogenies are of fundamental importance to all of comparative biology, including fields as diverse as conservation, forensics, agriculture, and medicine.
The Quantity and Quality of Phylogenetic Information
Modern genome sequencing techniques offer evolutionary biologists a wealth of data to use when inferring phylogenetic relationships. However, not all parts of the genome provide the same amount of phylogenetic information, nor is the information from different regions equally reliable. These challenges require new statistical approaches to quantifying information content and assessing the fit of the statistical models we use to interpret genome sequences. Our lab has a series of ongoing projects on these themes, including...
-
...developing new approaches to assessing model fit for phylogenomics.
-
...attempting to identify sets of genes that provide more reliable phylogenetic signal.
-

-
...asking whether model fit affects our ability to understand how many species exist.
-
...developing software for assessing phylogenetic model fit.
Modeling Molecular Evolution
In addition to providing unprecedented opportunities for understanding evolution, genomic data also pose new challenges. Our lab is interested in tailoring and extending phylogenetic methods to handle these challenges and take full advantage of our new sequencing capabilities. Some of these projects include...
-
...developing new models of sequence evolution that are tailored to specific types of genomic sequences (e.g., ultraconserved elements).
-
...using the information available in genomic databases as informed priors for Bayesian analyses.
-
...investigating site-specific patterns of rate shifts across phylogenies (heterotachy).
-
...whether non-random patterns of missing sequence data matter for phylogenetics.
Biodiversity, Speciation, and Comparative Analyses
Some of the most fundamental questions in evolutionary biology center around understanding how many species exist, what they are, how they formed, and how their characteristics (phenotypes) have evolved. We work collaboratively on a number of questions related to these goals, including...

-
...studying how the boundaries between frog species are maintained (or not).
-
...investigating how turtles relate to other tetrapod species.
-
...understanding the phylogenetic relationships of major squamate (lizard and snake) groups.
-
...exploring whether particular gene families affect rates of speciation and extinction.
-
...reconstructing the demographic history of species affected by habitat alteration.
Viral Evolution and Forensics
Viruses are particularly interesting to evolutionary biologists because they evolve so much more quickly than most other organisms. In addition, they are major forces affecting human health, global ecology, and the long-term evolutionary dynamics of their hosts. Our lab applies phylogenetic tools both to understand fundamental aspects of viral biology and to reconstruct their transmission patterns. Recently, we have collaborated with other labs to study...
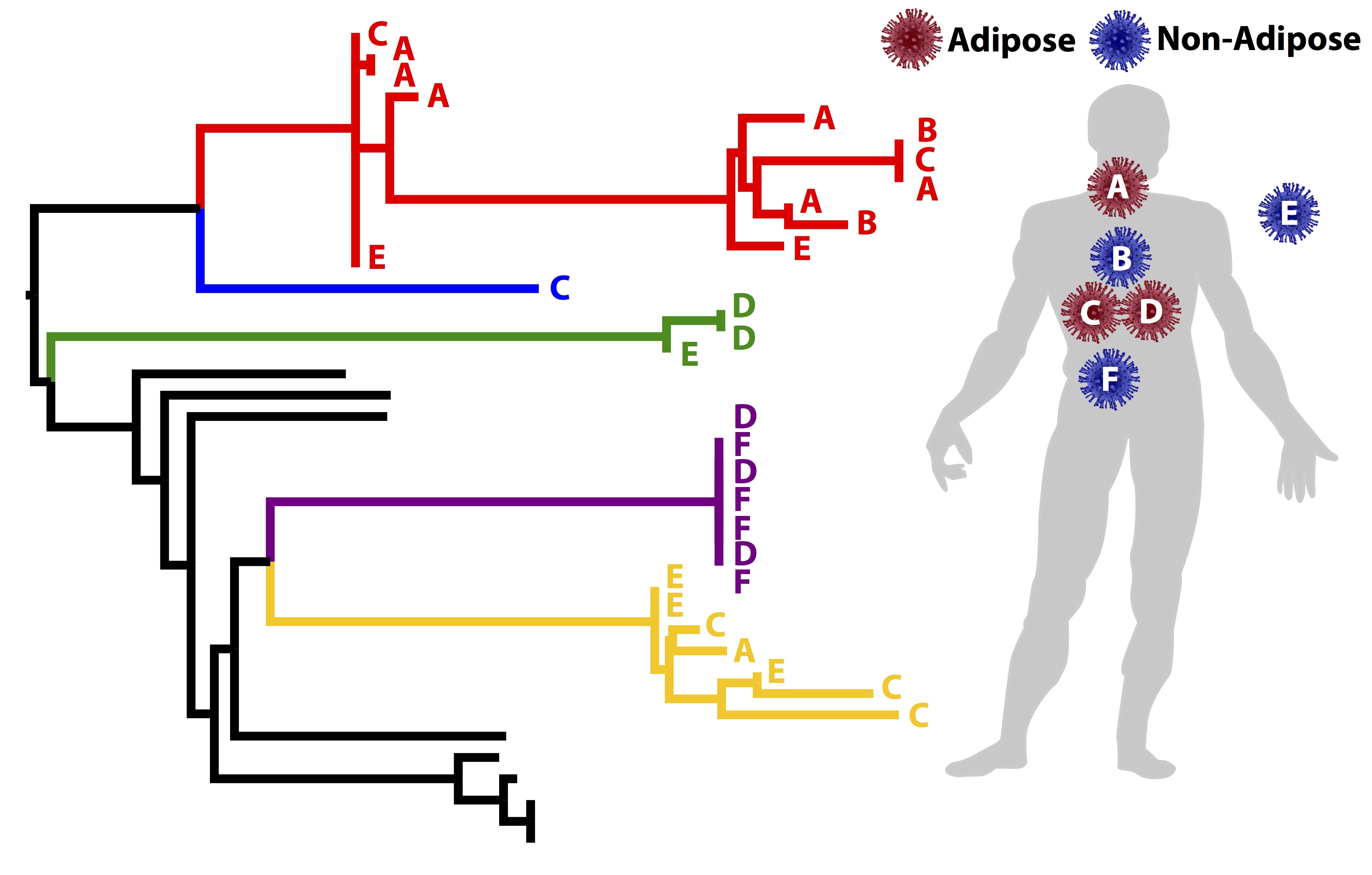
-
...how different parts of a particular protein (glycoprotein K) in alphaherpesviruses (like chicken pox) have evolved and how evolution can be used to understand protein function.